3 Figure 3
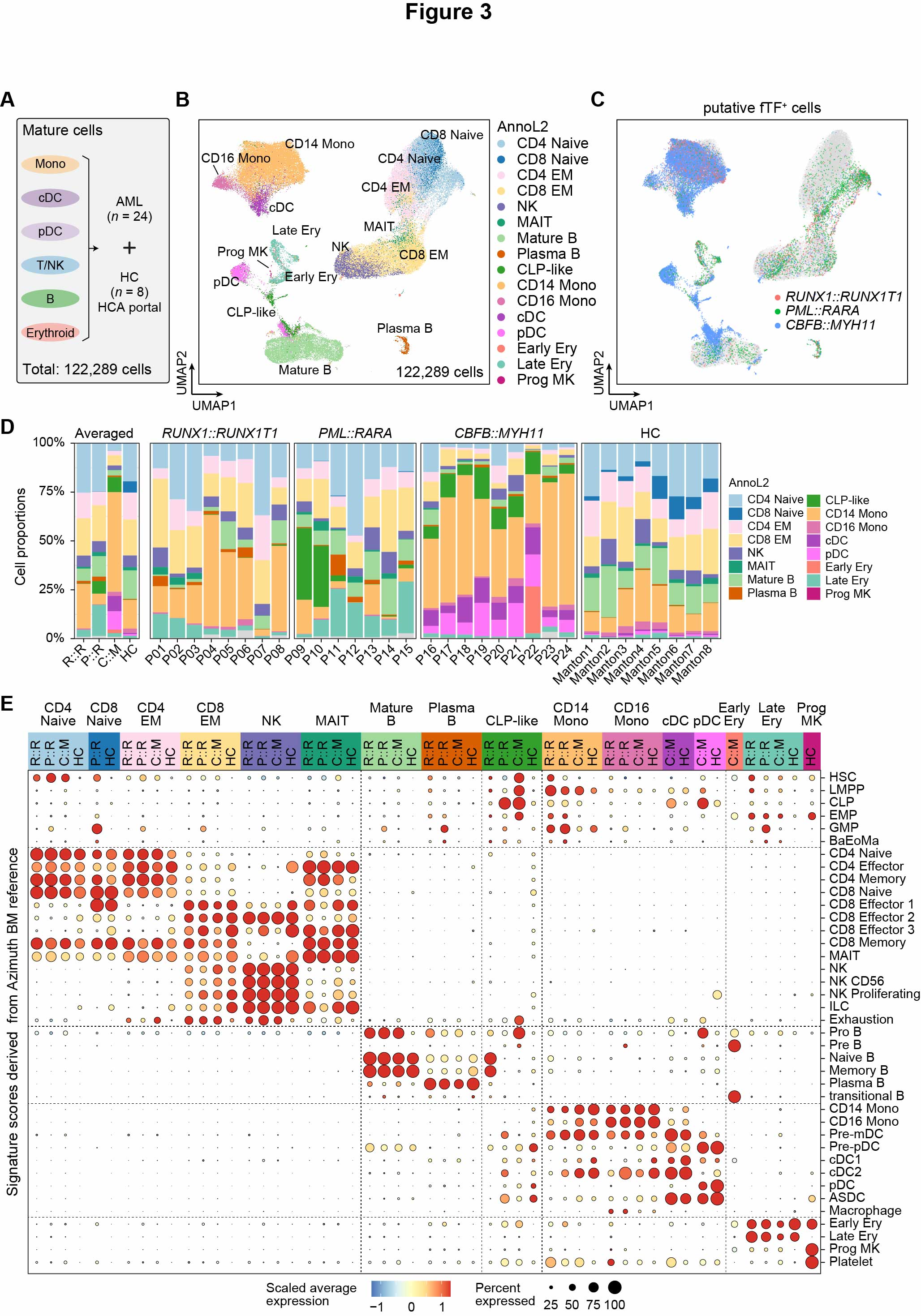 Fig. 3. Transcriptional aberrations in mature myeloid and lymphoid cells
Fig. 3. Transcriptional aberrations in mature myeloid and lymphoid cells
(A) Illustration of merged analysis of mature cells from AML BMs, using mature cells from 8 healthy BM samples as controls. (B) UMAP visualization of de novo clustering of mature cells from RUNX1::RUNX1T1, PML::RARA, CBFB::MYH11 AML and HC samples. The total number of sequenced cells is indicated in the bottom-right corner. (C) UMAP visualization showing the distribution of putative fTF+ cells: RUNX1::RUNX1T1 (red), PML::RARA (green), and CBFB::MYH11 (blue), with grey representing putative fTF- cells. (D) Bar charts depicting the relative fractions of 16 annotated cell types (annoL2) across different groups. The left panel shows the average fractions for each group, while the other panels present individual sample fractions. (E) Dot plots showing signature scores for gene sets representing distinct hematopoietic cell types, based on Azimuth BM cell type annotation level 2. R::R, RUNX1::RUNX1T1 AML; P::R, PML::RARA AML; C::M, CBFB::MYH11 AML.
3.1 (B) UMAP distribution
scAML.O.anno <- read_rds(paste0(in_dir, "Table1.3.scAML.O_harmony.anno.rds"))
anno_color.O <- c("#A6CEE3", "#1F78B4", "#FDDAEC", "#FEE090", "#7570B3", "#1B9E77",
"#A1D99B", "#D95F02", "#33A02C",
"#FDBF6F", "#DE77AE", "#AB47BC", "#FF77F8",
"#FB8072", "#73C8B4", "#C51B7D", "#D9D9D9")
anno_name.O <- c("CD4Naive", "CD8Naive", "CD4EM", "CD8EM", "NK", "MAIT",
"MatureB", "PlasmaB", "CLP-like",
"CD14Mono", "CD16Mono", "cDC", "pDC",
"EarlyE", "LateE", "ProgMK", "LowQual")
names(anno_color.O) <- anno_name.O
pdf(paste0(out_dir, "Fig3B.pdf"), width = 6, height = 4.5)
DimPlot(scAML.O.anno, reduction = "umap", group.by = "annoL2",
label = T, repel = T, raster = T, cols = anno_color.O, pt.size = 0.3)
dev.off()3.2 (C) Fusion distribution
scAML.O.anno.sub <- scAML.O.anno %>% subset(annoL2 != "LowQual")
res_pred <- read_rds(paste0(in_dir, "Table1.res_pred.351594.rds")) %>%
rownames_to_column("ID") %>% dplyr::select(ID, SS.final_fus_group)
df <- scAML.O.anno.sub@meta.data %>%
rownames_to_column("ID") %>%
left_join(., res_pred) %>%
mutate(fus_group2 = case_when(
(SS.final_fus_group == "Malignant" & FAB == "M2AE") ~ "M2AE",
(SS.final_fus_group == "Malignant" & FAB == "M3PR") ~ "M3PR",
(SS.final_fus_group == "Malignant" & FAB == "M4CM") ~ "M4CM",
(SS.final_fus_group == "Unclear") ~ "ANA")) %>%
left_join(., scAML.O.anno.sub@reductions$umap@cell.embeddings %>% data.frame() %>% rownames_to_column("ID")) %>%
column_to_rownames("ID")
df1 <- df %>% filter(SS.final_fus_group == "Unclear")
df2 <- df %>% filter(SS.final_fus_group != "Unclear") %>% sample_n(nrow(.))
pdf(paste0(out_dir, "Fig3C.pdf"), width = 4.7, height = 3.5)
rbind(df1, df2) %>%
mutate(mysize = ifelse((SS.final_fus_group == "Unclear"), 1, 2)) %>%
ggplot() +
ggrastr::rasterise(geom_point(aes_string(x = "umap_1", y = "umap_2", color = "fus_group2", size = "mysize"),
stroke = 0, alpha = 1), dpi = 300) +
scale_color_manual(values = c("#D9D9D9", "#F8766D", "#00BA38", "#619CFF")) +
scale_size_continuous(range = c(0.05, 0.2)) +
ggthemes::theme_few()
dev.off()3.3 (D) Bar charts
p1 <- plot_stat(scAML.O.anno, plot_type = "prop_fill", group_by = "FAB", pal_setup = anno_color.O) +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1))
p2 <- plot_stat(scAML.O.anno, plot_type = "prop_fill", group_by = "orig.ident", pal_setup = anno_color.O) +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1))
pdf(paste0(out_dir, "Fig3D.pdf"), width = 9, height = 4)
p1 + p2 + plot_layout(ncol = 2, widths = c(0.45, 3.8), guides = "collect")
dev.off()3.4 (E) Dot plots of signature scores
cd_features <- list(
HSC_score = c("CRHBP", "AVP", "MYCT1", "BEX1", "NPR3", "CRYGD", "MSRB3", "CD34", "NPDC1", "MLLT3"),
LMPP_score = c("AVP", "CRHBP", "C1QTNF4", "BEX1", "NPR3", "CD34", "NPW", "SMIM24", "CSF3R", "NPDC1"),
CLP_score = c("ACY3", "PRSS2", "C1QTNF4", "SPINK2", "SMIM24", "NREP", "CD34", "DNTT", "FLT3", "SPNS3"),
EMP_score = c("MYCT1", "CRHBP", "NPR3", "AVP", "GATA2", "HPGDS", "CYTL1", "CRYGD", "IGSF10", "PBX1"),
GMP_score = c("SERPINB10", "RNASE3", "MS4A3", "PRTN3", "ELANE", "AZU1", "CTSG", "RNASE2", "RETN", "NPW"),
BaEoMa_score = c("HDC", "MS4A2", "TPSAB1", "TPSB2", "CLC", "CPA3", "MS4A3", "IL1RL1", "PRG2", "HPGDS"),
Early_Eryth_score = c("CNRIP1", "GATA2", "ITGA2B", "TFR2", "GATA1", "KLF1", "CYTL1", "MAP7", "FSCN1", "APOC1"),
Late_Eryth_score = c("CTSE", "TSPO2", "IFIT1B", "TMEM56", "RHCE", "RHAG", "SPTA1", "ADD2", "EPCAM", "HBG1"),
Prog_Mk_score = c("CLEC1B", "SPX", "WFDC1", "ANXA3", "CMTM5", "SELP", "RBPMS2", "ARHGAP6", "GP9", "LTBP1"),
Platelet_score = c("RGS18", "C2orf88", "TMEM40", "GP9", "PF4", "PPBP", "DAB2", "SPARC", "RUFY1", "F13A1"),
CD14_Mono_score = c("FOLR3", "CLEC4E", "MCEMP1", "RBP7", "CDA", "FPR1", "CD300E", "C5AR1", "CD93", "APOBEC3A"),
CD16_Mono_score = c("LYPD2", "VMO1", "TPPP3", "C1QA", "C5AR1", "CD300E", "GPBAR1", "LILRA1", "HES4", "APOBEC3A"),
pre_mDC_score = c("ENHO", "CLEC10A", "RNASE2", "PLBD1", "FCER1A", "IGSF6", "MNDA", "SAMHD1", "ALDH2", "PAK1"),
pre_pDC_score = c("SCT", "SHD", "LILRA4", "LILRB4", "PTPRS", "TNNI2", "PLD4", "SPIB", "IRF8", "TNFRSF21"),
cDC1_score = c("IDO1", "XCR1", "TACSTD2", "CLEC9A", "WFDC21P", "CLNK", "CADM1", "DNASE1L3", "BATF3", "LGALS2"),
cDC2_score = c("ENHO", "CLEC10A", "CD1C", "IL13RA1", "PLBD1", "LGALS2", "CD33", "IGSF6", "IFI30", "FCER1A"),
pDC_score = c("ASIP", "CLEC4C", "SCAMP5", "PROC", "PTCRA", "SCT", "SHD", "PPM1J", "LILRA4", "LRRC26"),
ASDC_score = c("AXL", "SCT", "LILRA4", "PPP1R14A", "TGFBI", "UPK3A", "SCN9A", "CLEC10A", "PROC", "PLD4"),
Macrophage_score = c("SPIC", "FABP3", "CD5L", "CCL18", "C1QC", "C1QB", "FABP4", "C1QA", "APOE", "SELENOP"),
CD4_Naive_score = c("TSHZ2", "MAL", "CCR7", "CAMK4", "LEF1", "BCL11B", "FHIT", "RCAN3", "SH3YL1", "PIK3IP1"),
CD4_Effector_score = c("TNFRSF4", "AQP3", "TNFRSF25", "MAL", "IL7R", "TRAT1", "RORA", "FLT3LG", "KLRB1", "CD6"),
CD4_Memory_score = c("MAL", "TRAT1", "AP3M2", "CAMK4", "AQP3", "IL7R", "RCAN3", "BCL11B", "LEF1", "FLT3LG"),
CD8_Naive_score = c("NELL2", "CD8B", "S100B", "TRABD2A", "CCR7", "OXNAD1", "FLT3LG", "MAL", "LEF1", "CAMK4"),
CD8_Effector_1_score = c("CRTAM", "IFNG", "CD8A", "XCL1", "GZMK", "TIGIT", "CCL4L2", "CD8B", "CD160", "XCL2"),
CD8_Effector_2_score = c("ZNF683", "PRSS23", "KLRC2", "FCRL6", "S1PR5", "GZMH", "FGFBP2", "PATL2", "TRGC2", "CD3G"),
CD8_Effector_3_score = c("GZMK", "CD160", "CD8A", "KLRB1", "LAG3", "CCL4", "XCL2", "GZMM", "XCL1", "CCL3"),
CD8_Memory_score = c("CD8B", "CD8A", "GZMK", "IL7R", "BCL11B", "CD3E", "PIK3IP1", "CD6", "TRAC", "SPOCK2"),
T_Proliferating_score = c("RRM2", "MKI67", "BIRC5", "LCK", "SH2D1A", "CLSPN", "CDT1", "TK1", "TOP2A", "TRAC"),
MAIT_score = c("KLRB1", "IL7R", "TNFRSF25", "SPOCK2", "RORA", "NCR3", "GZMK", "KLRG1", "TRGC2", "CD6"),
NK_score = c("SH2D1B", "PRSS23", "S1PR5", "LAIR2", "KLRC1", "PTGDR", "FGFBP2", "FCRL6", "TBX21", "KLRC2"),
NK_CD56_score = c("SPTSSB", "NCAM1", "KLRC1", "XCL1", "IL18RAP", "XCL2", "IL2RB", "KLRF1", "TNFRSF18", "KLRD1"),
NK_Proliferating_score = c("GZMA", "GZMB", "TRDC", "PRF1", "TYMS", "FGFBP2", "KLRF1", "CD247", "GZMH", "CCL4"),
ILC_score = c("LDB2", "CD160", "XCL2", "KLRB1", "XCL1", "KLRF1", "KLRD1", "KLRC1", "TRDC", "GZMM"),
pro_B_score = c("CYGB", "UMODL1", "EBF1", "MME", "VPREB1", "DNTT", "IGLL1", "UHRF1", "BLNK", "AGPS"),
pre_B_score = c("NPY", "LCN6", "RAG2", "HMHB1", "ARPP21", "AKAP12", "RAG1", "C10orf10", "CYGB", "SLC8A1-AS1"),
Naive_B_score = c("FCRL2", "FCRL1", "ARHGAP24", "FCRL5", "FCER2", "FCRLA", "PAX5", "CD19", "HLA-DOB", "CD22"),
Memory_B_score = c("FCRL2", "TNFRSF13B", "ARHGAP24", "PNOC", "FCER2", "FCRLA", "BLK", "HLA-DOB", "MS4A1", "TNFRSF13C"),
Plasma_score = c("SDC1", "IGLC6", "IGLV6-57", "TNFRSF17", "IGLV3-1", "TNFRSF13B", "IGLC7", "JSRP1", "FCRL5", "IGKV1-5"),
transitional_B_score = c("BMP3", "CFAP73", "ARPP21", "IGLC5", "APBB2", "HRK", "DTX1", "AKAP12", "IGLC6", "MME"),
Exhaustion_score = c("CTLA4", "PDCD1", "TIGIT", "LAG3", "HAVCR2", "TOX"),
HLA1_score = c("HLA-A", "HLA-B", "HLA-C", "HLA-E", "HLA-F", "HLA-G"),
HLA2_score = c("HLA-DRA", "HLA-DRB", "HLA-DQA1", "HLA-DQA2", "HLA-DQB1", "HLA-DQB2", "HLA-DPA1", "HLA-DPA2", "HLA-DPB1", "HLA-DPB2", "HLA-DMA", "HLA-DMB", "HLA-DOA", "HLA-DOB", "HLA-DRB1", "HLA-DRB2", "HLA-DRB3", "HLA-DRB4", "HLA-DRB5", "HLA-DRB6", "HLA-DRB7", "HLA-DRB8", "HLA-DRB9")
)
idx_feature = rev(names(cd_features))
p <- DotPlot(object = scAML.O.anno, assay = "RNA", features = idx_feature, cols = "RdYlBu",
group.by = "FAB", split.by = "annoL2") +
geom_point(aes(size = pct.exp), shape = 21, stroke = 0.2) +
theme_bw() + theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) + coord_flip()
idx.sel <- scAML.O.anno@meta.data %>% group_by(FAB, annoL2) %>% summarise(N = n()) %>%
filter(N >= 10 & !annoL2 %in% "LowQual") %>% unite("id", c(FAB, annoL2)) %>% pull(id)
my_c <- unique(str_replace_all(idx.sel, "M2AE_|M3PR_|M4CM_|HC_BMMC_", ""))
####
df_raw <- p$data %>%
filter(id %in% idx.sel) %>%
mutate(features.plot = factor(features.plot,
levels = rev(names(cd_features)[c(1:6, 20:33, 40, 34:39, 11:19, 7:10, 41:42)]))) %>%
filter(features.plot %in% names(cd_features)[1:40]) %>%
mutate(FAB = str_extract(id, "M2AE|M3PR|M4CM|HC_BMMC"),
annoL2 = str_replace_all(id, "M2AE_|M3PR_|M4CM_|HC_BMMC_", "")) %>%
mutate(annoL2 = factor(annoL2, levels = anno_name.O[anno_name.O %in% my_c]))
p1 <- df_raw %>%
group_by(features.plot) %>%
mutate(avg.exp.scaled = as.numeric(scale(MinMax(log1p(avg.exp), -2, 50)))) %>% ungroup() %>%
mutate(avg.exp.scaled = MinMax(avg.exp.scaled, -1.3, 1.3)) %>%
ggplot() +
geom_point(aes(x = id, y = features.plot, fill = avg.exp.scaled, size = pct.exp), shape = 21, stroke = 0) +
facet_grid( ~ annoL2, scales = "free", space = "free") +
scale_fill_distiller(palette ="RdYlBu") +
theme_bw() + theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.5)) +
scale_radius(range = c(1, 6))
pdf(paste0(out_dir, "Fig3E.pdf"), width = 15, height = 9)
p1
dev.off()